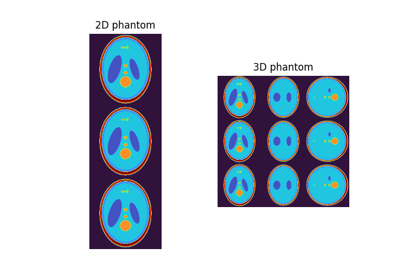
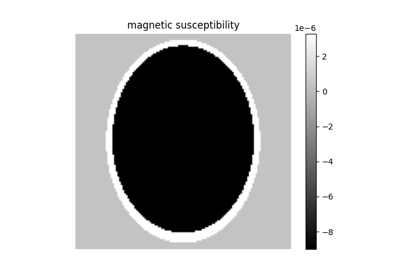
shepplogan_phantom#
- mrtwin.shepplogan_phantom(ndim, shape, model='single-pool', segtype='crisp', B0=1.5, cache=None, cache_dir=None)[source]#
Get SheppLogan phantom.
- Parameters:
ndim (int) – Number of spatial dimensions. If ndim == 2, use a single slice (central axial slice).
shape (int | Sequence[int]) – Shape of the output data. If int, assume isotropic matrix.
model (str, optional) –
String selecting one of the built-in tissue models. Valid entries are:
"single-pool": Single pool tissue model."mw-model": Myelin Water (MW) + Free Water (Intra-Extracellular, IEW)"mt-model": Macromolecular pool + Free Water (IEW + MW)"mwmt-model": Macromolecular pool + MW + IEW
The default is
"single-pool".segtype (str | bool, optional) – Phantom type. If it is a string (
"crisp") select crisp segmentation. If it isFalse, return a dense numeric phantom. The default iscrisp.B0 (float, optional) – Static field strength in [T]. The default is 1.5.
cache (bool | None, optional) – If
True, cache the phantom. The default isTruefor 3D phantoms andFalsefor single-slice 2D.cache_dir (CacheDirType, optional) – cache_directory for phantom caching. The default is
None(~/.cache/mrtwin).
- Returns:
Shepp-Logan phantom.
- Return type:
PhantomType
Examples
>>> import numpy as np >>> import matplotlib.pyplot as plt >>> from mrtwin import shepplogan_phantom
We can generate a dense single slice 2D phantom with a matrix size of
(200, 200)as:>>> phantom = shepplogan_phantom(ndim=2, shape=200, segtype=False)
Phantom T1 and T2 maps, can be accessed as:
>>> fig, ax = plt.subplots(2, 1)
>>> im1 = ax[0].imshow(phantom.T1, cmap="magma", vmax=1500) >>> ax[0].axis("off"), ax[0].set_title("T1 [ms]") >>> fig.colorbar(im1, ax=ax[0], fraction=0.046, pad=0.04)
>>> im2 = ax[1].imshow(phantom.T2, cmap="viridis", vmax=150) >>> ax[1].axis("off"), ax[1].set_title("T2 [ms]") >>> fig.colorbar(im, ax=ax[1], fraction=0.046, pad=0.04)