Note
Go to the end to download the full example code. or to run this example in your browser via Binder
Parameter Fitting#
This example shows how to use TorchSim to perform parameter inference.
We will build on the previous example. We will use numba, which can
be installed as pip install numba
We’ll generate an FSE dataset from IXI database. We will neglect encoding and assume single coil for this case.
import warnings
warnings.filterwarnings("ignore")
import os
import numpy as np
import torchio as tio
import torchsim
path = os.path.realpath("data")
ixi_dataset = tio.datasets.IXI(
path,
modalities=("PD", "T2"),
download=False,
)
# get subject 0
sample_subject = ixi_dataset[0]
M0 = sample_subject.PD.numpy().astype(np.float32).squeeze()[:, :, 60].T
T2w = sample_subject.T2.numpy().astype(np.float32).squeeze()[:, :, 60].T
sa = np.sin(np.deg2rad(8.0))
ta = np.tan(np.deg2rad(8.0))
T2 = -92.0 / np.log(T2w / M0)
T2 = np.nan_to_num(T2, neginf=0.0, posinf=0.0)
T2 = np.clip(T2, a_min=0.0, a_max=np.inf)
M0 = np.flip(M0)
T2 = np.flip(T2)
def simulate(T2, flip, ESP, device="cpu"):
# get ishape
ishape = T2.shape
output = torchsim.fse_sim(
flip=flip, ESP=ESP, T1=1000.0, T2=T2.flatten(), device=device
)
return abs(output.T.reshape(-1, *ishape)).numpy(force=True)
now generate the data
flip = 180.0 * np.ones(32, dtype=np.float32)
ESP = 5.0
device = "cpu"
# simulate acquisition
echo_series = M0 * simulate(T2, flip.copy(), ESP, device=device)
# display
img = np.concatenate((echo_series[0], echo_series[16], echo_series[-1]), axis=1)
import matplotlib.pyplot as plt
plt.imshow(abs(img), cmap="gray"), plt.axis("image"), plt.axis("off")
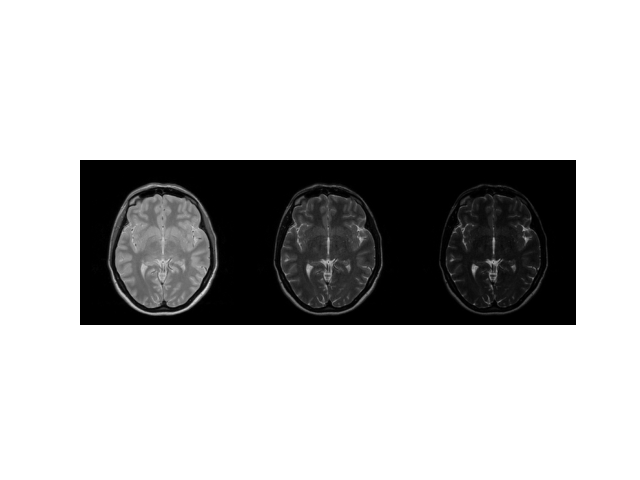
(<matplotlib.image.AxesImage object at 0x7ff48dfcead0>, (-0.5, 767.5, 255.5, -0.5), (-0.5, 767.5, 255.5, -0.5))
now, we want to implement a simple dictionary based inference algorithm. We first need a container to store the dictionary. We’ll use Python dataclasses for this:
from dataclasses import dataclass
@dataclass
class BlochDictionary:
atoms: np.ndarray
lookup_table: np.ndarray
labels: list
def __post_init__(self):
self.atoms = np.ascontiguousarray(self.atoms.transpose())
self.norm = np.linalg.norm(self.atoms, axis=0)
self.atoms = self.atoms / self.norm
self.lookup_table = np.ascontiguousarray(self.lookup_table.transpose())
self.labels = list(self.labels)
def to(self, device):
self.atoms = self.atoms.to(device)
self.norm = self.norm.to(device)
self.lookup_table = self.lookup_table.to(device)
return self
Now, we implement a simple exhaustive search algorithm. We’ll use Numba to parallelize it across different voxels:
import numba as nb
This is the main algorithm. We select matching entry using dot product as a cost function:
def _matching(signals, atoms, labels):
"""
performs pattern matching step.
"""
# preallocate
cost = np.zeros(signals.shape[0], dtype=np.complex64)
idx = np.zeros(signals.shape[0], dtype=int)
# do actual matching
_dot_search(signals, atoms, cost, idx)
return labels[:, idx], cost, idx
We need to implement the dot search:
@nb.njit(fastmath=True, parallel=True) # pragma: no cover
def _dot_search(time_series, dictionary, cost, idx):
for n in nb.prange(time_series.shape[0]):
for a in range(dictionary.shape[0]):
value = _dot_product(time_series[n], dictionary[a])
# keep maximum value
if np.abs(value) > np.abs(cost[n]):
cost[n] = value
idx[n] = a
Here, we implement a trivial dot product, compatible with numba:
@nb.njit(fastmath=True, cache=True) # pragma: no cover
def _dot_product(x, y):
z = 0.0
for n in range(x.shape[0]):
z += x[n] * y[n]
return z
We now create a wrapper to handle arbitrarily shaped inputs:
def matching(bloch_dict, time_series):
shape = time_series.shape[1:]
time_series = time_series.reshape((time_series.shape[0], np.prod(shape)))
time_series = np.ascontiguousarray(time_series.transpose().conj())
# get atoms
atoms = np.ascontiguousarray(bloch_dict.atoms.transpose())
labels = bloch_dict.lookup_table
# get quantitative maps and proton density
qmaps, cost, idx = _matching(time_series, atoms, labels)
qmaps = qmaps.reshape([qmaps.shape[0]] + list(shape))
qmaps = [qmap for qmap in qmaps]
m0 = (cost / bloch_dict.norm[idx]).reshape(shape)
return m0, dict(zip(bloch_dict.labels, qmaps))
We can assume the above code to be in a library. We now want to integrate it with our signal model from epg-torch-x. This can be done as:
import torch
def fse_fit(input, t2grid, flip, ESP, phases=None):
if isinstance(input, torch.Tensor):
istorch = True
device = input.device
input = input.numpy(force=True)
else:
istorch = False
# default
if phases is None:
phases = -np.ones_like(flip) * 90.0
# first build grid
t2lut = np.linspace(t2grid[0], t2grid[1], t2grid[2])
t1 = 1000.0
# build dictionary
atoms = torchsim.fse_sim(flip=flip, phases=phases, ESP=ESP, T1=t1, T2=t2lut).numpy(
force=True
)
blochdict = BlochDictionary(abs(atoms), t2lut[:, None], ["T2"])
# perform matching
m0, maps = matching(blochdict, input)
# here, we only have T2
t2map = maps["T2"]
# cast back
if istorch:
m0 = torch.as_tensor(m0, device=device)
t2map = torch.as_tensor(t2map, device=device)
return m0, t2map
Done! We can now try it:
M0rec, T2rec = fse_fit(echo_series.copy(), (1.0, 350.0, 1000), flip.copy(), ESP)
plt.subplot(2, 2, 1)
plt.imshow(T2, vmax=350.0), plt.axis("off"), plt.colorbar(), plt.title("true T2 [ms]")
plt.subplot(2, 2, 2)
plt.imshow(T2rec, vmax=350.0), plt.axis("off"), plt.colorbar(), plt.title(
"recon T2 [ms]"
)
plt.subplot(2, 2, 3)
plt.imshow(M0, cmap="gray"), plt.axis("off"), plt.colorbar(), plt.title("true M0")
plt.subplot(2, 2, 4)
plt.imshow(abs(M0rec), cmap="gray"), plt.axis("off"), plt.colorbar(), plt.title(
"recon M0"
)
![true T2 [ms], recon T2 [ms], true M0, recon M0](../../_images/sphx_glr_03-fitting_002.png)
(<matplotlib.image.AxesImage object at 0x7ff48cc0cac0>, (-0.5, 255.5, 255.5, -0.5), <matplotlib.colorbar.Colorbar object at 0x7ff48cc0f490>, Text(0.5, 1.0, 'recon M0'))
Total running time of the script: (0 minutes 2.642 seconds)