Note
Go to the end to download the full example code. or to run this example in your browser via Binder
Synthetic Data Generation#
This example shows how to use TorchSim to generate synthetic data.
We will use torchio and sigpy to get realistic ground truth maps and coil sensitivities. These can be installed as:
pip install torchio
pip install sigpy
We will use realistic maps from the IXI dataset,
downloaded using torchio:
import warnings
warnings.filterwarnings("ignore")
import os
import torchio as tio
path = os.path.realpath("data")
ixi_dataset = tio.datasets.IXI(
path,
modalities=("PD", "T2"),
download=False,
)
# get subject 0
sample_subject = ixi_dataset[0]
We will now extract an example slice and compute M0 and T2 maps to be used as simulation inputs.
import numpy as np
M0 = sample_subject.PD.numpy().astype(np.float32).squeeze()[:, :, 60].T
T2w = sample_subject.T2.numpy().astype(np.float32).squeeze()[:, :, 60].T
Compute T2 map:
Now, we can create our simulation function
Let’s use torchsim fse simulator
Assume a constant refocusing train
flip = 180.0 * np.ones(32, dtype=np.float32)
ESP = 5.0
device = "cpu"
# simulate acquisition
echo_series = M0 * simulate(T2, flip, ESP, device=device)
# display
img = np.concatenate((echo_series[0], echo_series[16], echo_series[-1]), axis=1)
import matplotlib.pyplot as plt
plt.imshow(abs(img), cmap="gray"), plt.axis("image"), plt.axis("off")
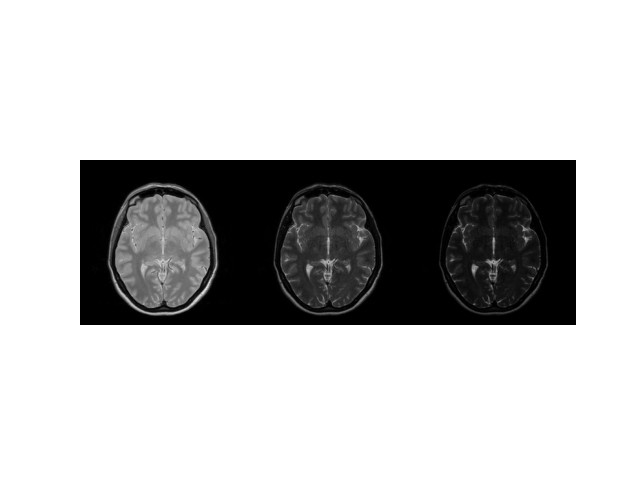
(<matplotlib.image.AxesImage object at 0x7ff497956920>, (-0.5, 767.5, 255.5, -0.5), (-0.5, 767.5, 255.5, -0.5))
Now, we want to add coil sensitivities. We will use Sigpy:
import sigpy.mri as smri
smaps = smri.birdcage_maps((8, *echo_series.shape[1:]))
We can simulate effects of coil by simple multiplication:
echo_series = smaps[:, None, ...] * echo_series
print(echo_series.shape)
(8, 32, 256, 256)
Now, we want to simulate k-space encoding. We will use a simple Poisson Cartesian encoding from Sigpy.
import sigpy as sp
mask = np.stack([smri.poisson(T2.shape, 32) for n in range(32)], axis=0)
ksp = mask * sp.fft(echo_series, axes=range(-2, 0))
plt.imshow(abs(ksp[0, 0]), vmax=50), plt.axis("image"), plt.axis("off"), plt.colorbar()
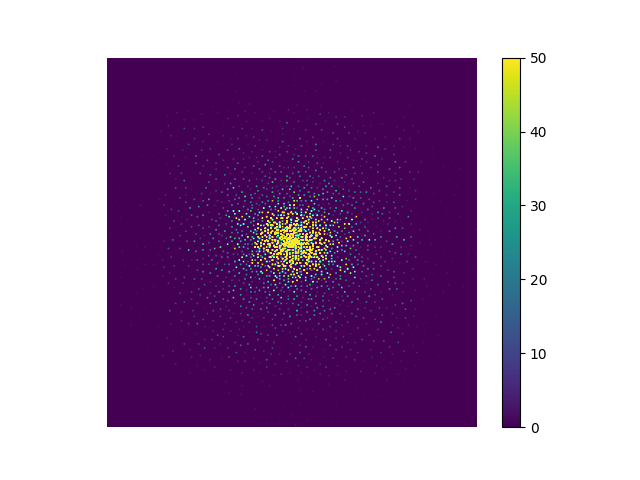
(<matplotlib.image.AxesImage object at 0x7ff49449a110>, (-0.5, 255.5, 255.5, -0.5), (-0.5, 255.5, 255.5, -0.5), <matplotlib.colorbar.Colorbar object at 0x7ff494143a90>)
Potentially, we could use Non-Cartesian sampling and include non-idealities
such as B0 accrual and T2* decay during readout using mri-nufft.
Now, we can wrap it up:
def generate_synth_data(M0, T2, flip, ESP, phases=None, ncoils=8, device="cpu"):
echo_series = M0 * simulate(T2, flip, ESP, device=device)
smaps = smri.birdcage_maps((ncoils, *echo_series.shape[1:]))
echo_series = smaps[:, None, ...] * echo_series
mask = np.stack(
[smri.poisson(T2.shape, len(flip)) for n in range(len(flip))], axis=0
)
return mask * sp.fft(echo_series, axes=range(-2, 0))
Reconstruction shows the effect of undersampling:
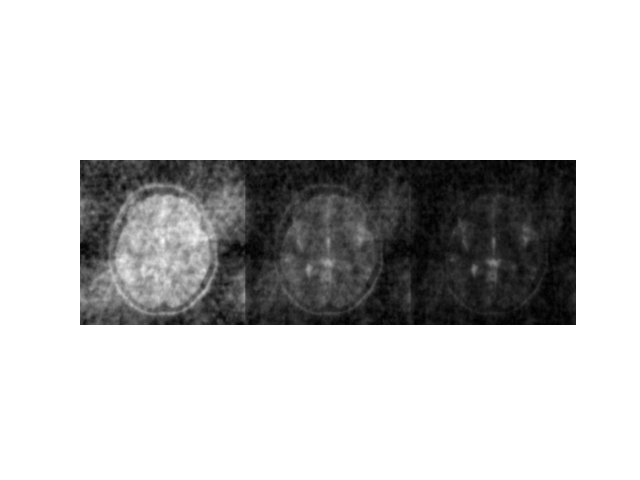
(<matplotlib.image.AxesImage object at 0x7ff48dfae440>, (-0.5, 767.5, 255.5, -0.5), (-0.5, 767.5, 255.5, -0.5))
This can be combined with data augmentation in torchio to generate synthetic datasets, such as in Synth-MOLED.
Total running time of the script: (0 minutes 12.080 seconds)